Generalization
To make DNA nano-cage transformable, we designed an adamantane-like structure equipped with several DNA hairpins which could work as fuelbetween edges. We utilized caDNAno to finish our design this nano-cage-like DNA origami with 5319 bps part of the M13mp18 chosen to beas the scaffold.
Our nano-cage has two conformations, one is the adamantane-like conformation (open state) and the other is the tetrahedron-like conformation (closed state). These two states have the potential to turn into each other by adding fuel or antifuel strands,respectively.
The nano-cage consists of 12 rods, 10 joints and 6 hairpin strands. Each section has its own function: the rods serve as basic holder, the joints provide the flexible area and the hairpin strands stimulate the whole structure to transform.
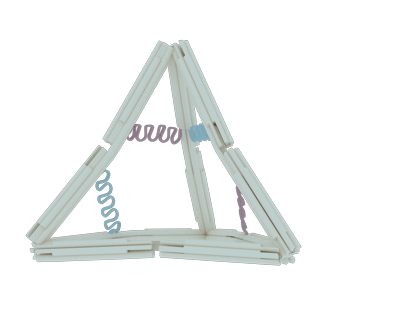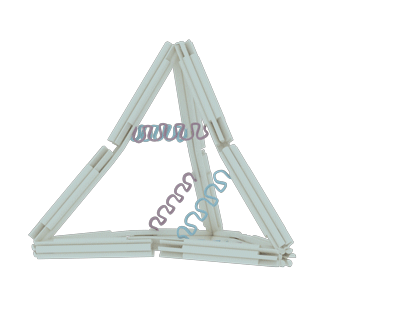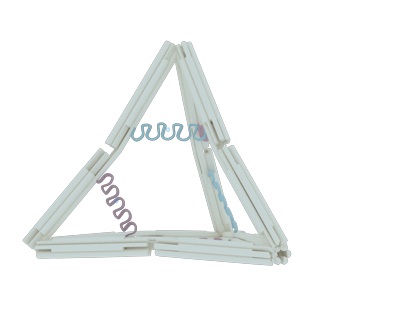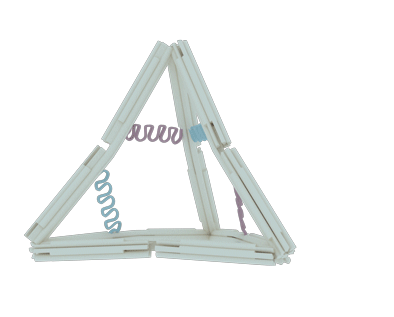

The rod
The rod was designed to be rigid enough to maintain the basic DNA nano-cage structure and to avoid disconnection in the shift of state. Since the natural DNA helix has a twist of 10.5 bps per turn, or 7 bps for 240 degrees of rotation, we insert 14 base pairs between each two adjacent crossovers, according to other documents’ origami design1-2, to make it twist naturally and therefore to gain the maximum stiffness (Figure 2). Each staple in the rod varies from 70 bps to 14 bps in length, but they all contribute to the construction of each rod in the form of six-helix helix-bundle honeycomb. In each rod, there are 8 staples, 8 crossovers and hence the rod’s length should be 63 bps or 21.4nm.
The joint
To endow our ball nano-cage with the potential to transform, we add flexible joints at both termini of each rods. The single strand of DNA is the best nominee for the joint due to its lower persistence length compared to the double strand of DNA. As the picture shows, each joint is a flexible area that contains only two or three single strands of scaffolds in the length of 12 bps. Among Ten ten joints, which three of them located aton four vertexes with three strands and the other six joints located at the middle of six edges with two strands, are incorporated into the nano-cage for rotational motion. The arrangement and freedom of the joints coupled with the stiff rods determine the reversible conformational change in our design.
The hairpin strands
After finishingFollowing the DNA nano-cage design, we had to deal with the energy problem: how to transform the state of nano-cage. We chose the DNA hairpin strands to solve the problem for its reconfigurability. We adjusted the state alterations by adding fuel strands to make the hairpins opened or by adding antifuel strands sequentially to make the hairpin closed. Along with the operation on hairpins, the nano-cage can also switch its forms between open and closed states.
| start | end | Hairpin sequence |
|---|---|---|
| 20[83] | 9[118] | AGCTGAAATCCAATGGAGACATAACGCGTAACAATCGCGAAAACGCGTAACAATC GCGAAAACGCGTAACAATCGCGAAAACGCGTAACAATCGCGAAAACGCGTAACAA TCGCGAAGACATGCGCCAATTCC |
| 8[118] | 2[112] | AGAATTATCAGACATAACCCCTAACAATGGGGAAAACCCCTAACAATGGGGAAAA CCCCTAACAATGGGGAAAACCCCTAACAATGGGGAAAACCCCTAACAATGGGGAA GACATGGCTAGCGAACCGCTCAA |
| 33[112] | 29[118] | AGAATGACAGACCAGGAGACATAACGCGTAACAATCGCGAAAACGCGTAACAATC GCGAAAACGCGTAACAATCGCGAAAACGCGTAACAATCGCGAAAACGCGTAACAA TCGCGAAGACATGCGCGGCACCA |
| 28[118] | 20[112] | CGTTAATTCAGACATAACCCCTAACAATGGGGAAAACCCCTAACAATGGGGAAAAC CCCTAACAATGGGGAAAACCCCTAACAATGGGGAAAACCCCTAACAATGGGGAAG ACATGGCTACCGTTCACCTTTA |
| 2[83] | 16[77] | CTGAACATTATCATGGAGACATAACGCGTAACAATCGCGAAAACGCGTAACAATCG CGAAAACGCGTAACAATCGCGAAAACGCGTAACAATCGCGAAAACGCGTAACAAT CGCGAAGACATGCGCTTTGCCT |
| 17[77] | 33[83] | GGAGGTTTCAGACATAACCCCTAACAATGGGGAAAACCCCTAACAATGGGGAAAA CCCCTAACAATGGGGAAAACCCCTAACAATGGGGAAAACCCCTAACAATGGGGAA GACATGGCTCGAACTATGAGATT |
To guarantee the ability of the nano-cage being spatially abundantfor protein encapsulation, we only stuck 6 hairpins on three ball’s faces while the other one being empty. This strategy will allow the encapsulation and release of CP easily when nuclestion site is incorporated. It’s sure been proven that 6 hairpins are adequate to drive the rods to move according to the AFM result.


The RNA loop
As we mentioned in introduction, the CP self-assembly begins at the (origin of assembly, )OAs region which we fixed into the nano-cage as CP detector. We designed an extended staple strand which reach out from an edge of the rod for hybridization with OAs. The sequence of OAs we picked up and the staple for fixing the OAs are shown in the table below.
| strand | sequence |
|---|---|
| RNA OAs | CCAUGGAACUUA CAGAAGAAGU CGUUGAUGAG UUCAUGGUAUAUAUA |
| DNA staple | GGAGGGAACAACTAAAGGAATTTTTTATACCGATAGTTGCCTTTCCGTATATATA |
The conformational change mechanism
When the fuel strands are added, the hybridization between hairpins and fuel strands results in the open state of hairpin and subsequently the tension reduction in apex joints, which benefits the increasing angle between rods. This transformation can be explained by the persistence length that quantify the stiffness of polymer. Informally, for pieces of the polymer shorter than the persistence length, the molecule behaves rather like a rigid rod/beam, while for pieces of the polymer longer than the persistence length, the molecule is much more flexible1. The persistence length of double stranded DNA from 100 bps to 160 bps is almost equal to the its physical length2, from which we assumed that the open hairpins are rigid enough to open the nano-cage. The AFM images have demonstrated that our assumption is correct.
In contrast, the complementary antifuel strands will remove the fuel strands from hairpins, leaving hairpins alone to form stem-loop structure that reduce the angle between rods. Considered the single stranded DNA (ssDNA) in joint are extremely flexible2 and hairpin has tendency to form secondary structure, it’s theoretically possible that the tension from the formation of stem-loop structure can overcome the bending energy from ssDNA, leading to the closed state of nano-cage consequently.
The protein encapsulation mechanism
In a longthe presence of TMV genome RNA, 34 CP units strongly interact with the OAs, forming a double-layered disk. Upon a transition of the double-disk into a helical “lockwasher”, the 5’ portion of the RNA is pulled “up” through the hole, allowing further CP disks to be concomitantly added to one end of the growing nanotube, intruding and coiling the RNA strand into the proteinaceous shell. Different from this mechanism of the RNA's 5’ end encapsidation, its 3’ end is thought to be packaged slowly by CP of lower oligomerization state and thus to remain accessible until the process is complete.

Because we picked only the partial OAs for the design, we believed the CP units will form the double-layered disk around the OAs and stop growing to become a helical “lockwasher” due to the fact that the length is not long enough for further elongation. When the nano-cage is open, CP units will aggregate assemble around OAs and fulfill the volume of nano-cage. When we added the antifuel, the nano-cage will turn into closed state, leading to the encapsulation of CP aggregate. Thus, the goal of catching TMV CP specifically is theoretically achieved.
REFERENCES
1. Kuzyk, A.; Yang, Y.; Duan, X.; Stoll, S.; Govorov, A. O.; Sugiyama, H.; Endo, M.; Liu, N., A light-driven three-dimensional plasmonic nanosystem that translates molecular motion into reversible chiroptical function. Nat Commun 2016, 7, 10591.
2. Douglas, S. M.; Dietz, H.; Liedl, T.; Hogberg, B.; Graf, F.; Shih, W. M., Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 2009, 459 (7245), 414-8.
3. Gutjahr, P.; Lipowsky, R.; Kierfeld, J., Persistence length of semiflexible polymers and bending rigidity renormalization. EPL (Europhysics Letters) 2006, 76 (6), 994.
4. Ouldridge, T. E.; Louis, A. A.; Doye, J. P., Structural, mechanical, and thermodynamic properties of a coarse-grained DNA model. The Journal of chemical physics 2011, 134 (8), 02B627.