Assembly and Characterization of DNA Origami
Asymmetric PCR For DNA Origami, the 5318 bp scaffold is amplified by the asymmetric Polymerase Chain Reaction (αPCR) from the M13 template. The Q5TMHigh-Fidelity DNA Polymerase (NEB M0491S) was used to guarantee the correctness of the sequence. Trying for times,the optimal PCR process was found and the optimal concentration ratio of sense primer to antisense primer for αPCR is 1:50, while the common ratios are between 1:50 to 1:100.(fig.1.b)The result of electrophoresis indicates that the scaffold was amplified successfully.
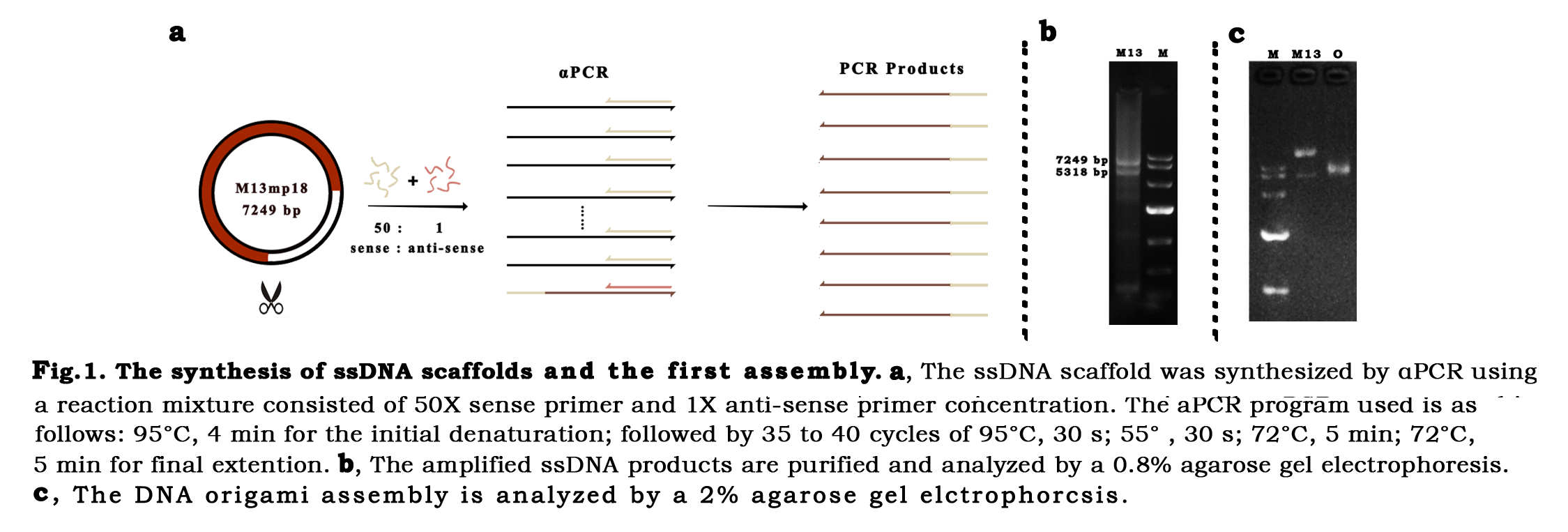
DNA self-assemblyOnce the scaffold was synthesized, the long single strand DNA was mixed with the short staple strands in a concentration ratio of 1:10. The one-pot reaction was performed through a long-playing annealing ramp, which allows each staple goes through its own annealing temperature, so that the scaffold and staples would pair correctly.The annealing products were first analyzed by electrophoresis. Confirming the assembly band (fig.1.c), the DNA nanostructure was characterized by AFM, DLS, TEM, successively.
AFM characterization As an efficient characterization, the Atomic Force Microscopy(AFM) discovered certain shapes of nanostructures which were preliminarily recognized to be the correct form that we designed(fig.2.a).
TEM characterizationTransmission Electron Microscopy (TEM) analysis was performed to reveal our nano-cage for imaging more clearly and obtaining the exact size. The DNA scaffold was designed to be folded into a tetrahedron-like frame with several six-helix-bundle nanotubes. The vertexes of the structure are unpaired DNA sequences, which make the nano-cage possible to transform between open and closed states. Long regulation strands are placed at the vertexes to provide the force to pull the cage close through formation of hairpin structures. After the one-pot annealing reaction, the hairpin bases paired and the cage turn into closed state. The micrographs (fig.2.b) confirmed that the cage assembled as expected. The size measured by the DLS is around 37.8 nm(fig.3),which is similar to the diameter measured on AFM images .
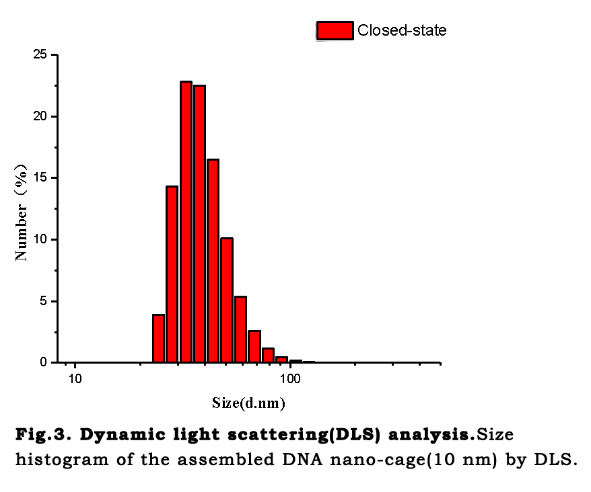
Mg2+ influenceTransmission Electron Microscopy (TEM) analysis was performed to reveal our nano-cage for
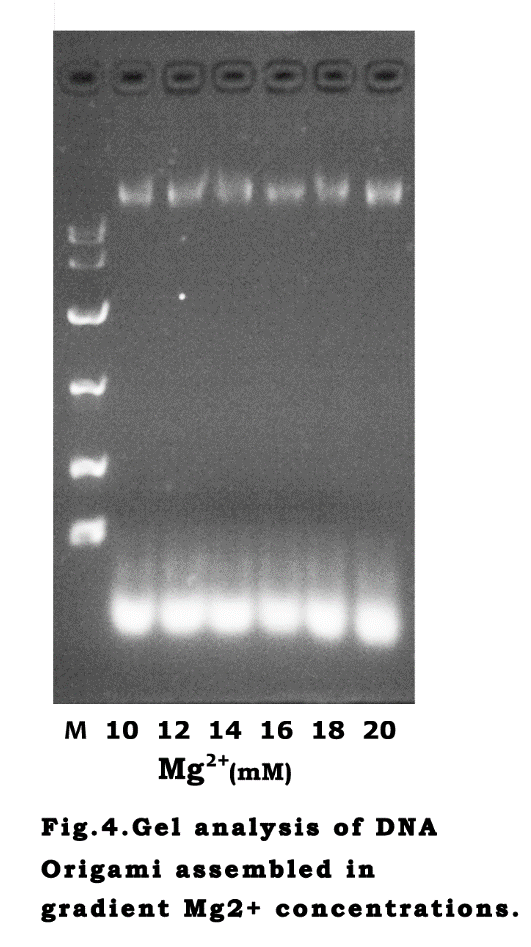
imaging more clearly and obtaining the exact size. The DNA scaffold was designed to be folded into a tetrahedron-like frame with several six-helix-bundle nanotubes. The vertexes of the structure are unpaired DNA sequences, which make the nano-cage possible to transform between open and closed states. Long regulation strands are placed at the vertexes to provide the force to pull the cage close through formation of hairpin structures. After the one-pot annealing reaction, the hairpin bases paired and the cage turn into closed state. The micrographs (fig.2.b) confirmed that the cage assembled as expected. The size measured by the DLS is around 37.8 nm(fig.3),which is similar to the diameter measured on AFM images .